Longest Common Subsequence Algorithm
The longest common subsequence problem is finding the longest sequence which exists in both the given strings.
But before we understand the problem, let us understand what the term subsequence is −
Let us consider a sequence S = <s1, s2, s3, s4, …,sn>. And another sequence Z = <z1, z2, z3, …,zm> over S is called a subsequence of S, if and only if it can be derived from S deletion of some elements. In simple words, a subsequence consists of consecutive elements that make up a small part in a sequence.
Common Subsequence
Suppose, X and Y are two sequences over a finite set of elements. We can say that Z is a common subsequence of X and Y, if Z is a subsequence of both X and Y.
Longest Common Subsequence
If a set of sequences are given, the longest common subsequence problem is to find a common subsequence of all the sequences that is of maximal length.
Naive Method
Let X be a sequence of length m and Y a sequence of length n. Check for every subsequence of X whether it is a subsequence of Y, and return the longest common subsequence found.
There are 2m subsequences of X. Testing sequences whether or not it is a subsequence of Y takes O(n) time. Thus, the naive algorithm would take O(n2m) time.
Longest Common Subsequence Algorithm
Let X=<x1,x2,x3....,xm> and Y=<y1,y2,y3....,ym> be the sequences. To compute the length of an element the following algorithm is used.
Step 1 − Construct an empty adjacency table with the size, n × m, where n = size of sequence X and m = size of sequence Y. The rows in the table represent the elements in sequence X and columns represent the elements in sequence Y.
Step 2 − The zeroeth rows and columns must be filled with zeroes. And the remaining values are filled in based on different cases, by maintaining a counter value.
Case 1 − If the counter encounters common element in both X and Y sequences, increment the counter by 1.
Case 2 − If the counter does not encounter common elements in X and Y sequences at T[i, j], find the maximum value between T[i-1, j] and T[i, j-1] to fill it in T[i, j].
Step 3 − Once the table is filled, backtrack from the last value in the table. Backtracking here is done by tracing the path where the counter incremented first.
Step 4 − The longest common subseqence obtained by noting the elements in the traced path.
Pseudocode
In this procedure, table C[m, n] is computed in row major order and another table B[m,n] is computed to construct optimal solution.
Algorithm: LCS-Length-Table-Formulation (X, Y)
m := length(X)
n := length(Y)
for i = 1 to m do
C[i, 0] := 0
for j = 1 to n do
C[0, j] := 0
for i = 1 to m do
for j = 1 to n do
if xi = yj
C[i, j] := C[i - 1, j - 1] + 1
B[i, j] := ‘D’
else
if C[i -1, j] ≥ C[i, j -1]
C[i, j] := C[i - 1, j] + 1
B[i, j] := ‘U’
else
C[i, j] := C[i, j - 1] + 1
B[i, j] := ‘L’
return C and B
Algorithm: Print-LCS (B, X, i, j) if i=0 and j=0 return if B[i, j] = ‘D’ Print-LCS(B, X, i-1, j-1) Print(xi) else if B[i, j] = ‘U’ Print-LCS(B, X, i-1, j) else Print-LCS(B, X, i, j-1)
This algorithm will print the longest common subsequence of X and Y.
Analysis
To populate the table, the outer for loop iterates m times and the inner for loop iterates n times. Hence, the complexity of the algorithm is O(m,n), where m and n are the length of two strings.
Example
In this example, we have two strings X=BACDB and Y=BDCB to find the longest common subsequence.
Following the algorithm, we need to calculate two tables 1 and 2.
Given n = length of X, m = length of Y
X = BDCB, Y = BACDB
Constructing the LCS Tables
In the table below, the zeroeth rows and columns are filled with zeroes. Remianing values are filled by incrementing and choosing the maximum values according to the algorithm.
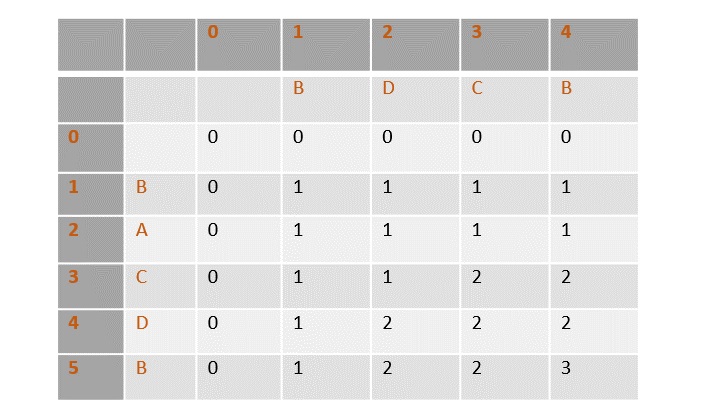
Once the values are filled, the path is traced back from the last value in the table at T[4, 5].
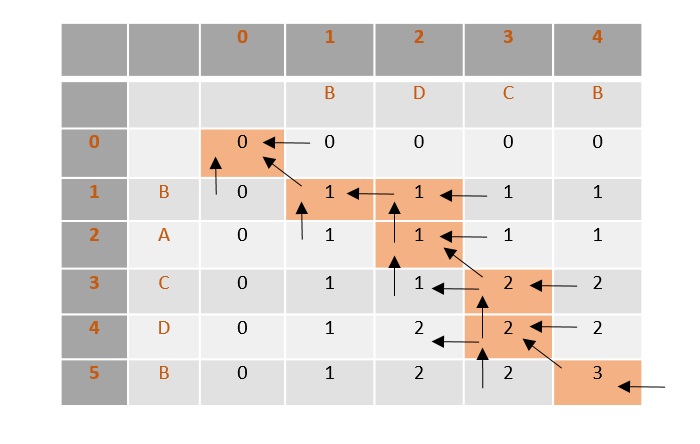
From the traced path, the longest common subsequence is found by choosing the values where the counter is first incremented.
In this example, the final count is 3 so the counter is incremented at 3 places, i.e., B, C, B. Therefore, the longest common subsequence of sequences X and Y is BCB.
Implementation
Following is the final implementation to find the Longest Common Subsequence using Dynamic Programming Approach −
#include <stdio.h>
#include <string.h>
int max(int a, int b);
int lcs(char* X, char* Y, int m, int n){
int L[m + 1][n + 1];
int i, j, index;
for (i = 0; i <= m; i++) {
for (j = 0; j <= n; j++) {
if (i == 0 || j == 0)
L[i][j] = 0;
else if (X[i - 1] == Y[j - 1]) {
L[i][j] = L[i - 1][j - 1] + 1;
} else
L[i][j] = max(L[i - 1][j], L[i][j - 1]);
}
}
index = L[m][n];
char LCS[index + 1];
LCS[index] = '\0';
i = m, j = n;
while (i > 0 && j > 0) {
if (X[i - 1] == Y[j - 1]) {
LCS[index - 1] = X[i - 1];
i--;
j--;
index--;
} else if (L[i - 1][j] > L[i][j - 1])
i--;
else
j--;
}
printf("LCS: %s
", LCS);
return L[m][n];
}
int max(int a, int b){
return (a > b) ? a : b;
}
int main(){
char X[] = "ABSDHS";
char Y[] = "ABDHSP";
int m = strlen(X);
int n = strlen(Y);
printf("Length of LCS is %d
", lcs(X, Y, m, n));
return 0;
}
Output
LCS: ABDHS Length of LCS is 5
#include <bits/stdc++.h>
using namespace std;
int max(int a, int b);
int lcs(char* X, char* Y, int m, int n){
int L[m + 1][n + 1];
int i, j, index;
for (i = 0; i <= m; i++) {
for (j = 0; j <= n; j++) {
if (i == 0 || j == 0)
L[i][j] = 0;
else if (X[i - 1] == Y[j - 1]) {
L[i][j] = L[i - 1][j - 1] + 1;
} else
L[i][j] = max(L[i - 1][j], L[i][j - 1]);
}
}
index = L[m][n];
char LCS[index + 1];
LCS[index] = '\0';
i = m, j = n;
while (i > 0 && j > 0) {
if (X[i - 1] == Y[j - 1]) {
LCS[index - 1] = X[i - 1];
i--;
j--;
index--;
} else if (L[i - 1][j] > L[i][j - 1])
i--;
else
j--;
}
printf("LCS: %s
", LCS);
return L[m][n];
}
int max(int a, int b){
return (a > b) ? a : b;
}
int main(){
char X[] = "ABSDHS";
char Y[] = "ABDHSP";
int m = strlen(X);
int n = strlen(Y);
printf("Length of LCS is %d
", lcs(X, Y, m, n));
return 0;
}
Output
LCS: ABDHS Length of LCS is 5
import java.util.*;
public class LCS_ALGO {
public static int max(int a, int b){
if( a > b){
return a;
}
else{
return b;
}
}
static int lcs(char arr1[], char arr2[], int m, int n) {
int[][] L = new int[m + 1][n + 1];
// Building the mtrix in bottom-up way
for (int i = 0; i <= m; i++) {
for (int j = 0; j <= n; j++) {
if (i == 0 || j == 0)
L[i][j] = 0;
else if (arr1[i - 1] == arr2[j - 1])
L[i][j] = L[i - 1][j - 1] + 1;
else
L[i][j] = max(L[i - 1][j], L[i][j - 1]);
}
}
int index = L[m][n];
int temp = index;
char[] lcs = new char[index + 1];
lcs[index] = '\0';
int i = m, j = n;
while (i > 0 && j > 0) {
if (arr1[i - 1] == arr2[j - 1]) {
lcs[index - 1] = arr1[i - 1];
i--;
j--;
index--;
}
else if (L[i - 1][j] > L[i][j - 1])
i--;
else
j--;
}
System.out.print("LCS: ");
for(i = 0; i<=temp; i++){
System.out.print(lcs[i]);
}
System.out.println();
return L[m][n];
}
public static void main(String[] args) {
String S1 = "ABSDHS";
String S2 = "ABDHSP";
char ch1[] = S1.toCharArray();
char ch2[] = S2.toCharArray();
int m = ch1.length;
int n = ch2.length;
System.out.println("Length of LCS is: " + lcs(ch1, ch2, m, n));
}
}
Output
LCS: ABDHS Length of LCS is: 5
def lcs(X, Y, m, n):
L = [[None]*(n+1) for a in range(m+1)]
for i in range(m+1):
for j in range(n+1):
if (i == 0 or j == 0):
L[i][j] = 0
elif (X[i - 1] == Y[j - 1]):
L[i][j] = L[i - 1][j - 1] + 1
else:
L[i][j] = max(L[i - 1][j], L[i][j - 1])
l = L[m][n]
LCS = [None] * (l)
a = m
b = n
while (a > 0 and b > 0):
if (X[a - 1] == Y[b - 1]):
LCS[l - 1] = X[a - 1]
a = a - 1
b = b - 1
l = l - 1
elif (L[a - 1][b] > L[a][b - 1]):
a = a - 1
else:
b = b - 1;
print("LCS is ")
print(LCS)
return L[m][n]
X = "ABSDHS"
Y = "ABDHSP"
m = len(X)
n = len(Y)
lc = lcs(X, Y, m, n)
print("Length of LCS is ")
print(lc)
Output
LCS is ['A', 'B', 'D', 'H', 'S'] Length of LCS is 5
Applications
The longest common subsequence problem is a classic computer science problem, the basis of data comparison programs such as the diff-utility, and has applications in bioinformatics. It is also widely used by revision control systems, such as SVN and Git, for reconciling multiple changes made to a revision-controlled collection of files.



